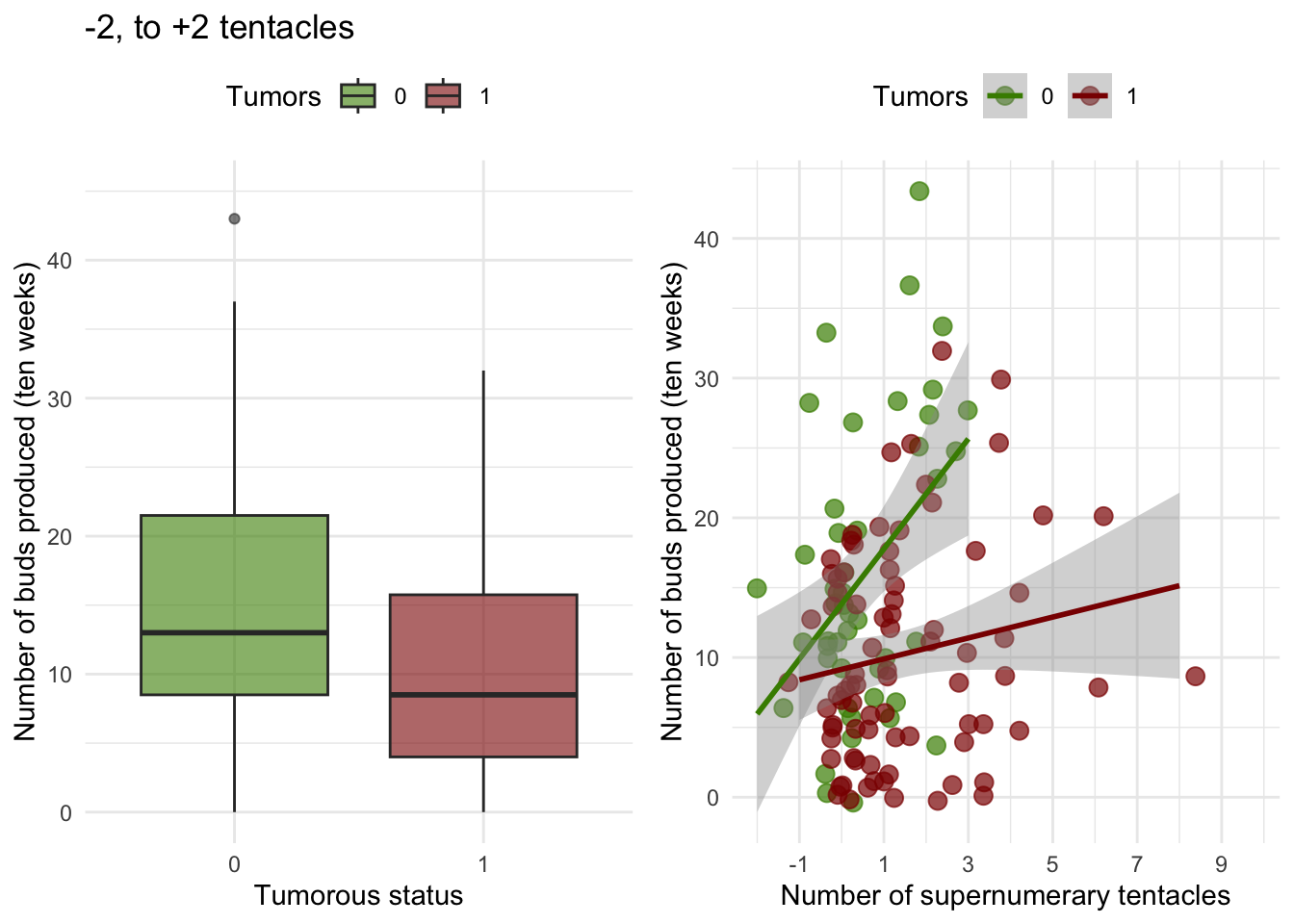
Supernumerary tentacles and fitness parameters of the host and the tumors
Budding and supernumerary tentacles
Transmissible tumors dataset
donor_transN <- subset(donor_trans, donor_trans$abnormalities == "Normal")
models <- glmulti(buds_10 ~ donor * donor_status * receiver * diff_maxR * Tumors,
data = donor_transN, level = 2, method = "h", crit = "aicc", fitfunction = "lm",
pl = FALSE)
tmp <- weightable(models)
tmp2 <- tmp[tmp$aicc <= min(tmp$aicc) + 2, ]
tmp2
best_rt1 <- lm(buds_10 ~ 1 + donor + donor_status + receiver + Tumors + diff_maxR +
receiver:donor_status + Tumors:receiver + Tumors:diff_maxR, data = donor_transN)
best_rt2 <- lm(buds_10 ~ 1 + donor + donor_status + receiver + Tumors + diff_maxR +
receiver:donor_status + Tumors:donor_status + Tumors:receiver, data = donor_transN)
best_rt3 <- lm(buds_10 ~ 1 + donor + donor_status + receiver + Tumors + diff_maxR +
receiver:donor_status + Tumors:donor_status + Tumors:receiver + Tumors:diff_maxR,
data = donor_transN)
best_rt4 <- lm(buds_10 ~ 1 + donor + donor_status + receiver + Tumors + diff_maxR +
donor_status:donor + receiver:donor_status + Tumors:receiver + Tumors:diff_maxR,
data = donor_transN)
best_rt5 <- lm(buds_10 ~ 1 + donor + donor_status + receiver + Tumors + diff_maxR +
receiver:donor_status + Tumors:receiver, data = donor_transN)
best_rt6 <- lm(buds_10 ~ 1 + donor + donor_status + receiver + Tumors + diff_maxR +
receiver:donor_status + Tumors:donor_status + Tumors:receiver + donor:diff_maxR,
data = donor_transN)
tab_model(best_rt1, best_rt2, best_rt3, best_rt4, best_rt5, best_rt6, show.intercept = F)
It appears that the appearance of tumors in certain receivers and the
interaction of recipient and donor status are the factors influencing
the most the number of buds produced. However, the number of tentacles
also seems to play a role that cannot be ruled out. To focus the
analysis on our parameters of interest, we will create groups of donors
and recipients to analyze the general relationship between tentacle
number and budding at the intra-group level.
Intra-group analysis
donor_transN$groupDR <- paste0(donor_transN$donor, donor_transN$donor_status, donor_transN$receiver)
best_rt10.4 <- lmer(buds_10 ~ 1 + diff_maxR * Tumors + (1 | groupDR), data = donor_transN)
best_rt10.3 <- lmer(buds_10 ~ 1 + diff_maxR + Tumors + (1 | groupDR), data = donor_transN)
best_rt10.2 <- lmer(buds_10 ~ 1 + diff_maxR + (1 | groupDR), data = donor_transN)
best_rt10.1 <- lmer(buds_10 ~ 1 + Tumors + (1 | groupDR), data = donor_transN)
best_rt10.0 <- lmer(buds_10 ~ 1 + (1 | groupDR), data = donor_transN)
AICc(best_rt10.4, best_rt10.3, best_rt10.2, best_rt10.1, best_rt10.0)## df AICc
## best_rt10.4 6 648.2747
## best_rt10.3 5 653.0875
## best_rt10.2 4 660.7359
## best_rt10.1 4 673.5175
## best_rt10.0 3 678.8187
Table of the results of the best fitted models (lower AICc+2)
| buds_10 | buds_10 | buds_10 | buds_10 | buds_10 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Predictors | Estimates | CI | p | Estimates | CI | p | Estimates | CI | p | Estimates | CI | p | Estimates | CI | p |
| diff maxR | 2.99 | 0.71 – 5.27 | 0.011 | 0.71 | -0.33 – 1.76 | 0.179 | 0.36 | -0.68 – 1.40 | 0.498 | ||||||
| Tumors [1] | -3.08 | -7.40 – 1.24 | 0.160 | -5.29 | -9.19 – -1.38 | 0.009 | -4.02 | -7.72 – -0.32 | 0.033 | ||||||
| diff maxR × Tumors [1] | -2.75 | -5.21 – -0.29 | 0.029 | ||||||||||||
| Random Effects | |||||||||||||||
| σ2 | 58.63 | 60.95 | 63.37 | 60.53 | 61.64 | ||||||||||
| τ00 | 19.45 groupDR | 21.64 groupDR | 33.02 groupDR | 24.18 groupDR | 32.80 groupDR | ||||||||||
| ICC | 0.25 | 0.26 | 0.34 | 0.29 | 0.35 | ||||||||||
| N | 8 groupDR | 8 groupDR | 8 groupDR | 8 groupDR | 8 groupDR | ||||||||||
| Observations | 92 | 92 | 92 | 95 | 95 | ||||||||||
| Marginal R2 / Conditional R2 | 0.115 / 0.335 | 0.072 / 0.315 | 0.004 / 0.345 | 0.044 / 0.317 | 0.000 / 0.347 | ||||||||||
simulateResiduals(best_rt10.4, plot = T)
Final model results:
tab_model(best_rt10.4, show.intercept = F)| buds_10 | |||
|---|---|---|---|
| Predictors | Estimates | CI | p |
| diff maxR | 2.99 | 0.71 – 5.27 | 0.011 |
| Tumors [1] | -3.08 | -7.40 – 1.24 | 0.160 |
| diff maxR × Tumors [1] | -2.75 | -5.21 – -0.29 | 0.029 |
| Random Effects | |||
| σ2 | 58.63 | ||
| τ00 groupDR | 19.45 | ||
| ICC | 0.25 | ||
| N groupDR | 8 | ||
| Observations | 92 | ||
| Marginal R2 / Conditional R2 | 0.115 / 0.335 | ||
If we look at the intra-group level, the number of supernumerary
tentacles and the presence of tumors interact significantly to explain
the budding rate.
When does this relationship establish ?
best_rt10 <- lmer(buds_10 ~ 1 + tenta_10 * Tumors + (1 | groupDR), data = donor_transN)
best_rt9 <- lmer(buds_9 ~ 1 + tenta_9 * Tumors + (1 | groupDR), data = donor_transN)
best_rt8 <- lmer(buds_8 ~ 1 + tenta_8 * Tumors + (1 | groupDR), data = donor_transN)
best_rt7 <- lmer(buds_7 ~ 1 + tenta_7 * Tumors + (1 | groupDR), data = donor_transN)
best_rt6 <- lmer(buds_6 ~ 1 + tenta_6 * Tumors + (1 | groupDR), data = donor_transN)
best_rt5 <- lmer(buds_5 ~ 1 + tenta_5 * Tumors + (1 | groupDR), data = donor_transN)
best_rt4 <- lmer(buds_4 ~ 1 + tenta_4 * Tumors + (1 | groupDR), data = donor_transN)
best_rt3 <- lmer(buds_3 ~ 1 + tenta_3 * Tumors + (1 | groupDR), data = donor_transN)
best_rt2 <- lmer(buds_2 ~ 1 + tenta_2 * Tumors + (1 | groupDR), data = donor_transN)
best_rt1 <- lmer(buds_1 ~ 1 + tenta_1 * Tumors + (1 | groupDR), data = donor_transN)
tab_model(best_rt10, best_rt9, best_rt8, best_rt7, best_rt6, best_rt5, best_rt4,
best_rt3, best_rt2, best_rt1, show.intercept = F)| buds_10 | buds_9 | buds_8 | buds_7 | buds_6 | buds_5 | buds_4 | buds_3 | buds_2 | buds_1 | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Predictors | Estimates | CI | p | Estimates | CI | p | Estimates | CI | p | Estimates | CI | p | Estimates | CI | p | Estimates | CI | p | Estimates | CI | p | Estimates | CI | p | Estimates | CI | p | Estimates | CI | p |
| tenta 10 | 4.87 | 1.98 – 7.75 | 0.001 | |||||||||||||||||||||||||||
| Tumors [1] | 20.45 | 3.48 – 37.42 | 0.019 | 11.30 | 1.36 – 21.24 | 0.026 | 10.62 | 0.07 – 21.18 | 0.049 | 15.78 | 6.99 – 24.58 | 0.001 | 5.23 | -3.13 – 13.58 | 0.218 | 3.78 | -1.89 – 9.46 | 0.189 | 3.65 | -0.86 – 8.15 | 0.112 | 0.53 | -3.37 – 4.42 | 0.789 | -0.12 | -2.25 – 2.00 | 0.908 | -0.66 | -1.88 – 0.56 | 0.289 |
| tenta 10 × Tumors [1] | -4.57 | -7.58 – -1.56 | 0.003 | |||||||||||||||||||||||||||
| tenta 9 | 2.93 | 1.38 – 4.49 | <0.001 | |||||||||||||||||||||||||||
| tenta 9 × Tumors [1] | -2.68 | -4.43 – -0.94 | 0.003 | |||||||||||||||||||||||||||
| tenta 8 | 2.62 | 0.96 – 4.28 | 0.002 | |||||||||||||||||||||||||||
| tenta 8 × Tumors [1] | -2.23 | -4.04 – -0.42 | 0.016 | |||||||||||||||||||||||||||
| tenta 7 | 3.30 | 1.93 – 4.68 | <0.001 | |||||||||||||||||||||||||||
| tenta 7 × Tumors [1] | -3.16 | -4.69 – -1.62 | <0.001 | |||||||||||||||||||||||||||
| tenta 6 | 1.45 | 0.16 – 2.75 | 0.028 | |||||||||||||||||||||||||||
| tenta 6 × Tumors [1] | -1.12 | -2.61 – 0.36 | 0.136 | |||||||||||||||||||||||||||
| tenta 5 | 0.91 | 0.05 – 1.77 | 0.038 | |||||||||||||||||||||||||||
| tenta 5 × Tumors [1] | -0.77 | -1.77 – 0.22 | 0.128 | |||||||||||||||||||||||||||
| tenta 4 | 0.90 | 0.27 – 1.53 | 0.006 | |||||||||||||||||||||||||||
| tenta 4 × Tumors [1] | -0.72 | -1.48 – 0.05 | 0.066 | |||||||||||||||||||||||||||
| tenta 3 | 0.37 | -0.21 – 0.95 | 0.210 | |||||||||||||||||||||||||||
| tenta 3 × Tumors [1] | -0.14 | -0.82 – 0.55 | 0.699 | |||||||||||||||||||||||||||
| tenta 2 | -0.03 | -0.33 – 0.28 | 0.861 | |||||||||||||||||||||||||||
| tenta 2 × Tumors [1] | 0.04 | -0.34 – 0.42 | 0.846 | |||||||||||||||||||||||||||
| tenta 1 | -0.05 | -0.22 – 0.12 | 0.575 | |||||||||||||||||||||||||||
| tenta 1 × Tumors [1] | 0.14 | -0.07 – 0.36 | 0.189 | |||||||||||||||||||||||||||
| Random Effects | ||||||||||||||||||||||||||||||
| σ2 | 56.37 | 46.86 | 41.98 | 29.99 | 24.69 | 15.82 | 9.96 | 4.84 | 1.62 | 0.37 | ||||||||||||||||||||
| τ00 | 15.35 groupDR | 19.27 groupDR | 14.62 groupDR | 6.05 groupDR | 6.05 groupDR | 3.66 groupDR | 1.37 groupDR | 0.94 groupDR | 0.40 groupDR | 0.08 groupDR | ||||||||||||||||||||
| ICC | 0.21 | 0.29 | 0.26 | 0.17 | 0.20 | 0.19 | 0.12 | 0.16 | 0.20 | 0.18 | ||||||||||||||||||||
| N | 8 groupDR | 8 groupDR | 8 groupDR | 8 groupDR | 8 groupDR | 8 groupDR | 8 groupDR | 8 groupDR | 8 groupDR | 8 groupDR | ||||||||||||||||||||
| Observations | 95 | 91 | 111 | 116 | 125 | 138 | 146 | 147 | 152 | 154 | ||||||||||||||||||||
| Marginal R2 / Conditional R2 | 0.149 / 0.331 | 0.126 / 0.381 | 0.079 / 0.317 | 0.164 / 0.304 | 0.040 / 0.229 | 0.031 / 0.213 | 0.053 / 0.168 | 0.018 / 0.177 | 0.001 / 0.201 | 0.024 / 0.204 | ||||||||||||||||||||
The relationship between the number of tentacles, the tumor presence,
and the budding seems to appear only after the third week, corresponding
to the establishment of the tumorous phenotype and the increase in the
number of tentacles.
Spontaneaous tumors dataset
donor_spontN <- subset(donor_spont, donor_spont$abnormalities == "Normal")
models <- glmulti(buds_10 ~ donor * donor_status * receiver * diff_maxR * Tumors,
data = donor_spontN, level = 2, method = "h", crit = "aicc", fitfunction = "lm",
pl = FALSE)
tmp <- weightable(models)
tmp2 <- tmp[tmp$aicc <= min(tmp$aicc) + 2, ]
tmp2
best_rt1 <- lm(buds_10 ~ 1 + donor + receiver + Tumors + diff_maxR + receiver:donor +
Tumors:donor + donor_status:diff_maxR, data = donor_spontN)
best_rt2 <- lm(buds_10 ~ 1 + donor + receiver + Tumors + diff_maxR + receiver:donor +
Tumors:donor + donor_status:diff_maxR + receiver:diff_maxR, data = donor_spontN)
best_rt3 <- lm(buds_10 ~ 1 + donor + receiver + diff_maxR + receiver:donor + donor:diff_maxR +
donor_status:diff_maxR, data = donor_spontN)
best_rt4 <- lm(buds_10 ~ 1 + donor + donor_status + receiver + Tumors + diff_maxR +
receiver:donor + Tumors:donor + donor_status:diff_maxR, data = donor_spontN)
best_rt5 <- lm(buds_10 ~ 1 + donor + donor_status + receiver + diff_maxR + receiver:donor +
donor:diff_maxR + donor_status:diff_maxR, data = donor_spontN)
best_rt6 <- lm(buds_10 ~ 1 + donor + receiver + Tumors + diff_maxR + receiver:donor +
Tumors:donor + donor_status:diff_maxR + Tumors:diff_maxR, data = donor_spontN)
best_rt7 <- lm(buds_10 ~ 1 + donor + receiver + Tumors + diff_maxR + receiver:donor +
Tumors:donor + donor_status:diff_maxR + Tumors:diff_maxR, data = donor_spontN)
best_rt8 <- lm(buds_10 ~ 1 + donor + donor_status + receiver + Tumors + diff_maxR +
donor_status:donor + receiver:donor + Tumors:donor + donor_status:diff_maxR,
data = donor_spontN)
best_rt9 <- lm(buds_10 ~ 1 + donor + receiver + Tumors + diff_maxR + receiver:donor +
Tumors:donor + donor:diff_maxR + donor_status:diff_maxR, data = donor_spontN)
tab_model(best_rt1, best_rt2, best_rt3, best_rt4, best_rt5, best_rt6, best_rt7, best_rt8,
best_rt9, show.intercept = F)
It seems that the appearance of tumors in certain donors and the
interaction of recipient and donor are the factors influencing the most
the number of buds. The number of tentacles also seems to play a role
when the donor was tumorous. To focus the analysis on the parameters of
interest, we will create groups of donors and recipients.
Intra group analysis
donor_spontN$groupDR <- as.factor(paste0(donor_spontN$donor, donor_spontN$donor_status,
donor_spontN$receiver))
summary(donor_spontN$groupDR)## MTNTSpB MTNTTV MTTSpB MTTTV SpBNTSpB SpBNTTV SpBTSpB SpBTTV
## 6 10 7 9 19 28 10 6best_rt10.4 <- lmer(buds_10 ~ 1 + diff_maxR * Tumors + (1 | groupDR), data = donor_spontN)
best_rt10.3 <- lmer(buds_10 ~ 1 + diff_maxR + Tumors + (1 | groupDR), data = donor_spontN)
best_rt10.2 <- lmer(buds_10 ~ 1 + diff_maxR + (1 | groupDR), data = donor_spontN)
best_rt10.1 <- lmer(buds_10 ~ 1 + Tumors + (1 | groupDR), data = donor_spontN)
best_rt10.0 <- lmer(buds_10 ~ 1 + (1 | groupDR), data = donor_spontN)
AICc(best_rt10.4, best_rt10.3, best_rt10.2, best_rt10.1, best_rt10.0)## df AICc
## best_rt10.4 6 373.7325
## best_rt10.3 5 376.3829
## best_rt10.2 4 377.8042
## best_rt10.1 4 399.9832
## best_rt10.0 3 400.6056Table of the results of the best fitted models (lower AICc+2)
| buds_10 | buds_10 | buds_10 | buds_10 | buds_10 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Predictors | Estimates | CI | p | Estimates | CI | p | Estimates | CI | p | Estimates | CI | p | Estimates | CI | p |
| diff maxR | -1.59 | -5.37 – 2.19 | 0.402 | 0.83 | -0.60 – 2.25 | 0.248 | 0.64 | -0.73 – 2.01 | 0.352 | ||||||
| Tumors [1] | -2.29 | -5.84 – 1.25 | 0.200 | -1.64 | -5.09 – 1.81 | 0.346 | -0.62 | -3.80 – 2.56 | 0.697 | ||||||
| diff maxR × Tumors [1] | 2.79 | -1.25 – 6.83 | 0.172 | ||||||||||||
| Random Effects | |||||||||||||||
| σ2 | 29.43 | 30.04 | 30.20 | 29.18 | 28.83 | ||||||||||
| τ00 | 20.67 groupDR | 20.23 groupDR | 18.91 groupDR | 20.26 groupDR | 19.60 groupDR | ||||||||||
| ICC | 0.41 | 0.40 | 0.39 | 0.41 | 0.40 | ||||||||||
| N | 8 groupDR | 8 groupDR | 8 groupDR | 8 groupDR | 8 groupDR | ||||||||||
| Observations | 58 | 58 | 58 | 62 | 62 | ||||||||||
| Marginal R2 / Conditional R2 | 0.040 / 0.436 | 0.021 / 0.415 | 0.010 / 0.391 | 0.002 / 0.411 | 0.000 / 0.405 | ||||||||||
tab_model(best_rt10.4, show.intercept = F)| buds_10 | |||
|---|---|---|---|
| Predictors | Estimates | CI | p |
| diff maxR | -1.59 | -5.37 – 2.19 | 0.402 |
| Tumors [1] | -2.29 | -5.84 – 1.25 | 0.200 |
| diff maxR × Tumors [1] | 2.79 | -1.25 – 6.83 | 0.172 |
| Random Effects | |||
| σ2 | 29.43 | ||
| τ00 groupDR | 20.67 | ||
| ICC | 0.41 | ||
| N groupDR | 8 | ||
| Observations | 58 | ||
| Marginal R2 / Conditional R2 | 0.040 / 0.436 | ||
If we look at the intra-group level, there is no clear relationship,
which is quite expected given the absence of an increased number of
tentacles when the donor was a spontaneous tumor.
When does this relationship establish ?
best_rt10 <- lmer(buds_10 ~ 1 + tenta_10 * Tumors + (1 | groupDR), data = donor_spontN)
best_rt9 <- lmer(buds_9 ~ 1 + tenta_9 * Tumors + (1 | groupDR), data = donor_spontN)
best_rt8 <- lmer(buds_8 ~ 1 + tenta_8 * Tumors + (1 | groupDR), data = donor_spontN)
best_rt7 <- lmer(buds_7 ~ 1 + tenta_7 * Tumors + (1 | groupDR), data = donor_spontN)
best_rt6 <- lmer(buds_6 ~ 1 + tenta_6 * Tumors + (1 | groupDR), data = donor_spontN)
best_rt5 <- lmer(buds_5 ~ 1 + tenta_5 * Tumors + (1 | groupDR), data = donor_spontN)
best_rt4 <- lmer(buds_4 ~ 1 + tenta_4 * Tumors + (1 | groupDR), data = donor_spontN)
best_rt3 <- lmer(buds_3 ~ 1 + tenta_3 * Tumors + (1 | groupDR), data = donor_spontN)
best_rt2 <- lmer(buds_2 ~ 1 + tenta_2 * Tumors + (1 | groupDR), data = donor_spontN)
best_rt1 <- lmer(buds_1 ~ 1 + tenta_1 * Tumors + (1 | groupDR), data = donor_spontN)
tab_model(best_rt10, best_rt9, best_rt8, best_rt7, best_rt6, best_rt5, best_rt4,
best_rt3, best_rt2, best_rt1, show.intercept = F)| buds_10 | buds_9 | buds_8 | buds_7 | buds_6 | buds_5 | buds_4 | buds_3 | buds_2 | buds_1 | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Predictors | Estimates | CI | p | Estimates | CI | p | Estimates | CI | p | Estimates | CI | p | Estimates | CI | p | Estimates | CI | p | Estimates | CI | p | Estimates | CI | p | Estimates | CI | p | Estimates | CI | p |
| tenta 10 | 0.94 | -2.33 – 4.21 | 0.568 | |||||||||||||||||||||||||||
| Tumors [1] | -6.84 | -23.33 – 9.64 | 0.409 | -6.17 | -17.01 – 4.67 | 0.259 | -3.51 | -15.38 – 8.36 | 0.557 | -1.71 | -11.55 – 8.14 | 0.730 | -6.97 | -15.84 – 1.91 | 0.122 | -2.05 | -8.39 – 4.28 | 0.521 | -1.84 | -5.94 – 2.26 | 0.375 | -1.02 | -4.99 – 2.95 | 0.610 | -1.22 | -3.57 – 1.13 | 0.305 | -0.23 | -1.66 – 1.20 | 0.748 |
| tenta 10 × Tumors [1] | 0.90 | -2.48 – 4.27 | 0.597 | |||||||||||||||||||||||||||
| tenta 9 | 0.04 | -1.86 – 1.94 | 0.967 | |||||||||||||||||||||||||||
| tenta 9 × Tumors [1] | 1.14 | -1.02 – 3.30 | 0.294 | |||||||||||||||||||||||||||
| tenta 8 | 0.33 | -1.77 – 2.44 | 0.752 | |||||||||||||||||||||||||||
| tenta 8 × Tumors [1] | 0.70 | -1.59 – 2.99 | 0.543 | |||||||||||||||||||||||||||
| tenta 7 | 0.83 | -0.72 – 2.37 | 0.289 | |||||||||||||||||||||||||||
| tenta 7 × Tumors [1] | 0.33 | -1.60 – 2.27 | 0.730 | |||||||||||||||||||||||||||
| tenta 6 | -0.26 | -1.91 – 1.39 | 0.756 | |||||||||||||||||||||||||||
| tenta 6 × Tumors [1] | 1.48 | -0.31 – 3.26 | 0.103 | |||||||||||||||||||||||||||
| tenta 5 | 0.50 | -0.64 – 1.64 | 0.384 | |||||||||||||||||||||||||||
| tenta 5 × Tumors [1] | 0.39 | -0.87 – 1.65 | 0.537 | |||||||||||||||||||||||||||
| tenta 4 | 0.44 | -0.02 – 0.90 | 0.060 | |||||||||||||||||||||||||||
| tenta 4 × Tumors [1] | 0.33 | -0.45 – 1.10 | 0.403 | |||||||||||||||||||||||||||
| tenta 3 | 0.28 | -0.40 – 0.95 | 0.414 | |||||||||||||||||||||||||||
| tenta 3 × Tumors [1] | 0.18 | -0.59 – 0.95 | 0.645 | |||||||||||||||||||||||||||
| tenta 2 | -0.05 | -0.40 – 0.29 | 0.765 | |||||||||||||||||||||||||||
| tenta 2 × Tumors [1] | 0.26 | -0.19 – 0.71 | 0.254 | |||||||||||||||||||||||||||
| tenta 1 | 0.05 | -0.14 – 0.24 | 0.592 | |||||||||||||||||||||||||||
| tenta 1 × Tumors [1] | 0.07 | -0.19 – 0.34 | 0.578 | |||||||||||||||||||||||||||
| Random Effects | ||||||||||||||||||||||||||||||
| σ2 | 24.80 | 29.06 | 22.96 | 17.56 | 12.25 | 8.53 | 5.15 | 3.59 | 1.28 | 0.33 | ||||||||||||||||||||
| τ00 | 18.39 groupDR | 13.96 groupDR | 11.59 groupDR | 5.69 groupDR | 4.76 groupDR | 3.65 groupDR | 1.81 groupDR | 0.86 groupDR | 0.23 groupDR | 0.03 groupDR | ||||||||||||||||||||
| ICC | 0.43 | 0.32 | 0.34 | 0.24 | 0.28 | 0.30 | 0.26 | 0.19 | 0.15 | 0.08 | ||||||||||||||||||||
| N | 8 groupDR | 8 groupDR | 8 groupDR | 8 groupDR | 8 groupDR | 8 groupDR | 8 groupDR | 8 groupDR | 8 groupDR | 8 groupDR | ||||||||||||||||||||
| Observations | 62 | 58 | 66 | 67 | 72 | 83 | 91 | 92 | 94 | 95 | ||||||||||||||||||||
| Marginal R2 / Conditional R2 | 0.114 / 0.491 | 0.060 / 0.365 | 0.049 / 0.368 | 0.060 / 0.290 | 0.092 / 0.346 | 0.062 / 0.343 | 0.077 / 0.317 | 0.049 / 0.234 | 0.021 / 0.170 | 0.042 / 0.117 | ||||||||||||||||||||
This is coherent; no relationship is observed due to not enough
variations.
Global Dataset: Spontaneous and Transmissible Tumors Together
Intra group analysis
data_1$groupDR <- as.factor(paste0(data_1$donor, data_1$donor_status, data_1$recipient))## Warning: Unknown or uninitialised column: `recipient`.summary(data_1$groupDR)## MTNT MTT RobNT RobT SpB_spontT SpBNT SpBT
## 17 16 37 36 20 56 46data_1N <- subset(data_1, data_1$abnormalities == "Normal")
best_rt10.4 <- lmer(buds_10 ~ 1 + diff_maxR * Tumors + (1 | groupDR), data = donor_transN)
best_rt10.3 <- lmer(buds_10 ~ 1 + diff_maxR + Tumors + (1 | groupDR), data = donor_transN)
best_rt10.2 <- lmer(buds_10 ~ 1 + diff_maxR + (1 | groupDR), data = donor_transN)
best_rt10.1 <- lmer(buds_10 ~ 1 + Tumors + (1 | groupDR), data = donor_transN)
best_rt10.0 <- lmer(buds_10 ~ 1 + Tumors + (1 | groupDR), data = donor_transN)
AICc(best_rt10.4, best_rt10.3, best_rt10.2, best_rt10.1, best_rt10.0)## df AICc
## best_rt10.4 6 648.2747
## best_rt10.3 5 653.0875
## best_rt10.2 4 660.7359
## best_rt10.1 4 673.5175
## best_rt10.0 4 673.5175Table of the results of the best fitted models (lower AICc+2)
| buds_10 | |||
|---|---|---|---|
| Predictors | Estimates | CI | p |
| diff maxR | 2.99 | 0.71 – 5.27 | 0.011 |
| Tumors [1] | -3.08 | -7.40 – 1.24 | 0.160 |
| diff maxR × Tumors [1] | -2.75 | -5.21 – -0.29 | 0.029 |
| Random Effects | |||
| σ2 | 58.63 | ||
| τ00 groupDR | 19.45 | ||
| ICC | 0.25 | ||
| N groupDR | 8 | ||
| Observations | 92 | ||
| Marginal R2 / Conditional R2 | 0.115 / 0.335 | ||
The combination of increased number of tentacles and tumor presence
explains a significant part of the budding rate experienced
intra-group.
When does this relationship establish ?
| buds_10 | buds_9 | buds_8 | buds_7 | buds_6 | buds_5 | buds_4 | buds_3 | buds_2 | buds_1 | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Predictors | Estimates | CI | p | Estimates | CI | p | Estimates | CI | p | Estimates | CI | p | Estimates | CI | p | Estimates | CI | p | Estimates | CI | p | Estimates | CI | p | Estimates | CI | p | Estimates | CI | p |
| tenta 10 | 4.81 | 2.40 – 7.23 | <0.001 | |||||||||||||||||||||||||||
| Tumors [1] | 17.13 | 3.43 – 30.82 | 0.015 | 7.40 | -1.82 – 16.62 | 0.115 | 8.33 | -0.81 – 17.47 | 0.074 | 12.69 | 4.99 – 20.39 | 0.001 | 1.33 | -5.89 – 8.56 | 0.716 | 2.54 | -2.55 – 7.63 | 0.326 | 1.97 | -1.77 – 5.71 | 0.300 | 0.65 | -2.75 – 4.05 | 0.706 | 0.16 | -1.81 – 2.13 | 0.873 | -0.46 | -1.66 – 0.73 | 0.447 |
| tenta 10 × Tumors [1] | -4.03 | -6.56 – -1.50 | 0.002 | |||||||||||||||||||||||||||
| tenta 9 | 2.76 | 1.25 – 4.26 | <0.001 | |||||||||||||||||||||||||||
| tenta 9 × Tumors [1] | -1.97 | -3.64 – -0.30 | 0.021 | |||||||||||||||||||||||||||
| tenta 8 | 2.63 | 1.11 – 4.15 | 0.001 | |||||||||||||||||||||||||||
| tenta 8 × Tumors [1] | -1.90 | -3.53 – -0.27 | 0.023 | |||||||||||||||||||||||||||
| tenta 7 | 3.32 | 2.08 – 4.55 | <0.001 | |||||||||||||||||||||||||||
| tenta 7 × Tumors [1] | -2.71 | -4.10 – -1.33 | <0.001 | |||||||||||||||||||||||||||
| tenta 6 | 1.46 | 0.31 – 2.61 | 0.013 | |||||||||||||||||||||||||||
| tenta 6 × Tumors [1] | -0.46 | -1.78 – 0.85 | 0.488 | |||||||||||||||||||||||||||
| tenta 5 | 1.27 | 0.49 – 2.05 | 0.002 | |||||||||||||||||||||||||||
| tenta 5 × Tumors [1] | -0.60 | -1.52 – 0.31 | 0.196 | |||||||||||||||||||||||||||
| tenta 4 | 0.93 | 0.41 – 1.45 | 0.001 | |||||||||||||||||||||||||||
| tenta 4 × Tumors [1] | -0.48 | -1.13 – 0.18 | 0.152 | |||||||||||||||||||||||||||
| tenta 3 | 0.60 | 0.08 – 1.12 | 0.025 | |||||||||||||||||||||||||||
| tenta 3 × Tumors [1] | -0.19 | -0.80 – 0.43 | 0.549 | |||||||||||||||||||||||||||
| tenta 2 | 0.16 | -0.12 – 0.43 | 0.262 | |||||||||||||||||||||||||||
| tenta 2 × Tumors [1] | -0.02 | -0.38 – 0.33 | 0.893 | |||||||||||||||||||||||||||
| tenta 1 | 0.03 | -0.13 – 0.20 | 0.688 | |||||||||||||||||||||||||||
| tenta 1 × Tumors [1] | 0.10 | -0.11 – 0.32 | 0.343 | |||||||||||||||||||||||||||
| Random Effects | ||||||||||||||||||||||||||||||
| σ2 | 52.28 | 48.67 | 40.33 | 28.46 | 23.60 | 15.43 | 9.58 | 5.04 | 1.71 | 0.43 | ||||||||||||||||||||
| τ00 | 11.56 groupDR | 13.03 groupDR | 9.59 groupDR | 4.52 groupDR | 3.50 groupDR | 2.08 groupDR | 0.63 groupDR | 0.27 groupDR | 0.12 groupDR | 0.01 groupDR | ||||||||||||||||||||
| ICC | 0.18 | 0.21 | 0.19 | 0.14 | 0.13 | 0.12 | 0.06 | 0.05 | 0.07 | 0.03 | ||||||||||||||||||||
| N | 7 groupDR | 7 groupDR | 7 groupDR | 7 groupDR | 7 groupDR | 7 groupDR | 7 groupDR | 7 groupDR | 7 groupDR | 7 groupDR | ||||||||||||||||||||
| Observations | 132 | 122 | 148 | 153 | 165 | 181 | 192 | 194 | 199 | 202 | ||||||||||||||||||||
| Marginal R2 / Conditional R2 | 0.156 / 0.309 | 0.114 / 0.301 | 0.097 / 0.270 | 0.167 / 0.281 | 0.079 / 0.198 | 0.083 / 0.192 | 0.084 / 0.140 | 0.055 / 0.103 | 0.013 / 0.077 | 0.030 / 0.059 | ||||||||||||||||||||
Again, very coherent results; the relationship starts when the tumorous
phenotype, including supernumerary tentacles, is expressed.
## df AICc
## best_rt10.1 6 909.2623
## best_rt10.2 5 918.7976
## best_rt10.3 4 926.3033
## best_rt10.4 4 924.1592
## best_rt10.5 2 943.7884| buds_10 | buds_10 | |||||
|---|---|---|---|---|---|---|
| Predictors | Estimates | CI | p | Estimates | CI | p |
| tenta 10 | 4.81 | 2.40 – 7.23 | <0.001 | |||
| Tumors [1] | 17.13 | 3.43 – 30.82 | 0.015 | -3.34 | -6.35 – -0.32 | 0.030 |
| tenta 10 × Tumors [1] | -4.03 | -6.56 – -1.50 | 0.002 | |||
| Random Effects | ||||||
| σ2 | 52.28 | 57.58 | ||||
| τ00 | 11.56 groupDR | 20.30 groupDR | ||||
| ICC | 0.18 | 0.26 | ||||
| N | 7 groupDR | 7 groupDR | ||||
| Observations | 132 | 132 | ||||
| Marginal R2 / Conditional R2 | 0.156 / 0.309 | 0.033 / 0.285 | ||||
The relationship between the direct number of tentacles at week 10 and
budding rate is even stronger; however, we prefer to keep a consistent
indicator in the final analysis.
Data visualisation
## ID Manipulator donor donor_status donor_tentacle
## 2 : 1 Justine: 62 MT :30 NT:85 Min. : 4.000
## 4 : 1 Océane :109 Rob :55 T :86 1st Qu.: 6.000
## 5 : 1 SpB :71 Median : 6.000
## 6 : 1 SpB_spont:15 Mean : 7.327
## 13 : 1 3rd Qu.: 8.000
## 14 : 1 Max. :18.000
## (Other):165
## receiver receiver_tentacle lot date_draft abnormalities
## SpB:80 Min. :3.000 22 : 18 05/04 :21 Excluded: 0
## TV :91 1st Qu.:5.000 18 : 11 12/04 :18 Lost : 0
## Median :6.000 11 : 10 29/03 :18 Normal :171
## Mean :5.637 12 : 10 08/04 :15 Sick : 0
## 3rd Qu.:6.000 16 : 10 04/02 :13
## Max. :8.000 19 : 10 01/02 :11
## (Other):102 (Other):75
## dateT Tumors dateD Death tenta_1 tenta_2
## Min. : 6.00 0:79 Min. :11.00 0:108 Min. :3.000 Min. :2.000
## 1st Qu.:20.00 1:92 1st Qu.:34.00 1: 63 1st Qu.:5.000 1st Qu.:5.000
## Median :28.00 Median :41.00 Median :6.000 Median :6.000
## Mean :32.46 Mean :42.94 Mean :5.561 Mean :5.476
## 3rd Qu.:42.00 3rd Qu.:56.00 3rd Qu.:6.000 3rd Qu.:6.000
## Max. :70.00 Max. :70.00 Max. :8.000 Max. :8.000
## NA's :80 NA's :108 NA's :3
## tenta_3 tenta_4 tenta_5 tenta_6
## Min. :0.000 Min. :0.000 Min. :0.000 Min. :1.000
## 1st Qu.:5.000 1st Qu.:5.000 1st Qu.:5.000 1st Qu.:5.000
## Median :6.000 Median :5.000 Median :5.000 Median :5.000
## Mean :5.417 Mean :5.385 Mean :5.377 Mean :5.331
## 3rd Qu.:6.000 3rd Qu.:6.000 3rd Qu.:6.000 3rd Qu.:6.000
## Max. :8.000 Max. :8.000 Max. :8.000 Max. :8.000
## NA's :8 NA's :10 NA's :20 NA's :35
## tenta_7 tenta_8 tenta_9 tenta_10
## Min. :2.000 Min. :0.000 Min. :0.000 Min. :0.000
## 1st Qu.:5.000 1st Qu.:5.000 1st Qu.:4.000 1st Qu.:5.000
## Median :6.000 Median :5.000 Median :5.000 Median :5.000
## Mean :5.464 Mean :5.342 Mean :5.296 Mean :5.415
## 3rd Qu.:6.000 3rd Qu.:6.000 3rd Qu.:6.000 3rd Qu.:6.000
## Max. :8.000 Max. :8.000 Max. :8.000 Max. :9.000
## NA's :46 NA's :51 NA's :73 NA's :65
## buds_1 buds_2 buds_3 buds_4
## Min. :0.0000 Min. :0.0000 Min. : 0.000 Min. : 0.000
## 1st Qu.:0.0000 1st Qu.:0.0000 1st Qu.: 0.000 1st Qu.: 1.000
## Median :0.0000 Median :0.0000 Median : 1.000 Median : 3.000
## Mean :0.3099 Mean :0.7976 Mean : 1.969 Mean : 3.559
## 3rd Qu.:0.0000 3rd Qu.:1.0000 3rd Qu.: 3.000 3rd Qu.: 6.000
## Max. :4.0000 Max. :6.0000 Max. :11.000 Max. :15.000
## NA's :3 NA's :8 NA's :10
## buds_5 buds_6 buds_7 buds_8
## Min. : 0.000 Min. : 0.000 Min. : 0.000 Min. : 0.00
## 1st Qu.: 2.000 1st Qu.: 3.000 1st Qu.: 4.000 1st Qu.: 4.00
## Median : 5.000 Median : 6.000 Median : 8.000 Median : 9.00
## Mean : 5.477 Mean : 7.199 Mean : 8.683 Mean :10.04
## 3rd Qu.: 9.000 3rd Qu.:10.250 3rd Qu.:13.000 3rd Qu.:14.25
## Max. :18.000 Max. :22.000 Max. :27.000 Max. :33.00
## NA's :20 NA's :35 NA's :45 NA's :51
## buds_9 buds_10 Taille tumeur avant congelation
## Min. : 0.00 Min. : 0.00 Min. :0.000
## 1st Qu.: 4.00 1st Qu.: 6.00 1st Qu.:0.000
## Median :10.00 Median :11.00 Median :1.000
## Mean :10.77 Mean :12.26 Mean :1.819
## 3rd Qu.:16.00 3rd Qu.:16.75 3rd Qu.:3.000
## Max. :39.00 Max. :43.00 Max. :6.000
## NA's :73 NA's :65 NA's :66
## Date congelation tenta_max diff_max diff_maxR
## Length:171 Min. :3.000 Min. :0.0000 Min. :-2.0000
## Class :character 1st Qu.:5.000 1st Qu.:0.0000 1st Qu.: 0.0000
## Mode :character Median :6.000 Median :0.0000 Median : 0.0000
## Mean :6.029 Mean :0.4678 Mean : 0.3918
## 3rd Qu.:7.000 3rd Qu.:1.0000 3rd Qu.: 1.0000
## Max. :9.000 Max. :3.0000 Max. : 2.0000
##
## groupDR
## MTNT :15
## MTT :15
## RobNT :29
## RobT :26
## SpB_spontT:15
## SpBNT :41
## SpBT :30## `geom_smooth()` using formula = 'y ~ x'